October 18, 2021
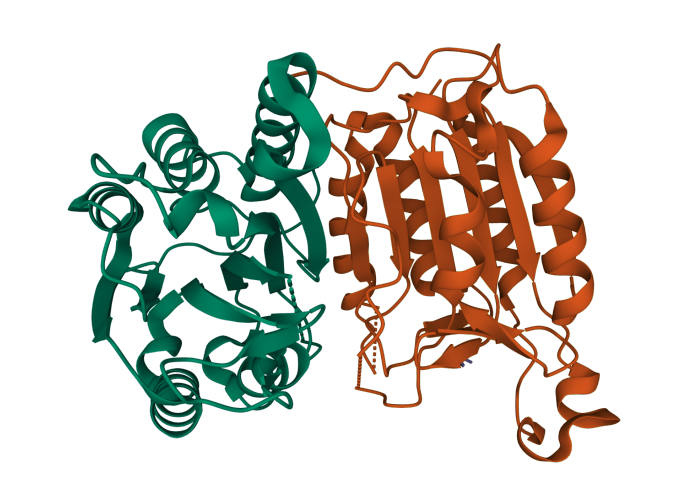
A Mol* (Sehnal et al. (2021)) visualization of interacting protein chains (PDB ID: 3H11).
A Mizzou Engineering team has released new software that will allow computers to automatically predict protein interactions. At the heart of the DeepInteract system is the Geometric Transformer, an algorithm specifically designed for learning from 3D protein shapes, which in turn helps predict interactions between proteins at an atomic level.

Morehead
Understanding how proteins interact and function together could help scientists design new proteins for specific purposes. Ultimately, that could accelerate the development of new prescription drugs, alternatives to fossil fuels, and other biological discoveries, said Alex Morehead, a PhD student in computer science who is leading the research.
“This is really exciting work,” he said. “The Geometric Transformer is a new idea that hasn’t been done before. That’s exciting, to push the boundaries of deep learning and help Mizzou become a leader in this field. It’s exciting and the first of many projects to come.”
Proteins are the basic building blocks of life. They begin as strings of amino acids that fold into three-dimensional shapes. Scientists have spent decades trying to develop a way to predict an individual protein’s structure.
Jianlin “Jack” Cheng, William and Nancy Thompson Distinguished Professor of electrical engineering and computer science, was among the first to apply deep learning to the problem, developing a model that predicts protein structures by measuring the distance of its specific amino acids. Last year, a Google-based company largely solved the protein prediction problem using an advanced deep learning approach.
DeepInteract builds upon that work. Instead of predicting a single protein’s shape, Morehead and Cheng — along with PhD student Chen Chen — looked at pairs of proteins, or binary complexes, to predict how proteins interact with each other when binding together.
“Shape is very important,” Morehead said. “If you don’t know the shape, it’s hard to completely understand what a protein is doing when it binds with another protein. When proteins bind together, they can change shape, and in such cases, we may expect their overall function to change as well.”

Cheng
To design and develop the Geometric Transformer, Morehead reviewed similar deep learning algorithms designed for learning from 3D objects. He then trained the model using 15,000 unique pairs of interacting proteins from DIPS-Plus, a database of interacting protein structures previously curated by Morehead, Chen, and Cheng earlier this year.
DeepInteract uses geometric deep learning — or machine learning designed to learn from objects with underlying geometries — to predict which atoms in a pair of proteins will interact with one another when the two proteins bind together.
“Proteins are made up of atoms, and you can connect the atoms together based on distance or other metrics such as nearest neighbors,” Morehead said. “Once you do that, you have three-dimensional renderings, and once you define that geometry, you have a lot of rich information from which your algorithm can learn. We then want to use whatever the machine learned from that geometry to predict atomic interactions between proteins. That’s DeepInteract in a nutshell.”
Morehead and Cheng have released a preprint of a paper currently under review, a common strategy in the ever-changing discipline of artificial intelligence. The team has also made the open-source software available on GitHub so others can reproduce the work.
“Others can use the instructions we provided or use what we created to predict new interactions,” Morehead said. “The key goal was to make something useful and let others build on top of that work.”