July 12, 2022

A Mizzou research team has developed a two-part system that accurately detects cancerous mutations in a single molecule of genetic material (DNA).
But wait, there’s more. Biomedical and Biological Engineering Professor Li-Qun “Andrew” Gu and his collaborators also believe his nanopore machine could have the ability to read digital data stored on DNA. The team recently filed a patent for the work.
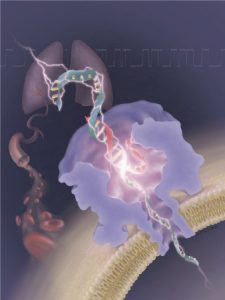
In the medical application, researchers devised a more accurate way to detect molecular mutations by capturing individual genetic markers. This can help doctors not only predict cancer risks but also determine the best treatment options for patients.
“A doctor wants to know the genetic point of mutation, but the problem is that even in late-stage cancer, not all of the cells have the same mutations,” Gu said. “Only a percentage of cells change to a tumor. In early cancer detection, maybe one out of a thousand cells becomes tumorous. So, this utilizes technology to detect the 0.1 percent of cells that have mutated to cancer and are susceptible to a targeted therapeutic drug.”
The patented system has two components. On one side, Kent Gates, a professor of chemistry, developed a tiny probe that, when mixed with the genetic material from a tumor sample/biopsy, identifies certain sequences of DNA that indicate a mutation. The probe then permanently latches on to that molecule.
“Our probe is covalent, which means it bites down on the sequence and doesn’t let go,” said Gates, who is also associate chair of chemistry in the College of Arts and Science. “That’s different from the way others work, as they are mostly weak and reversible attachments that can fall apart during analysis.”
Then, Gu’s nanopore device “catches” the molecules that have been tagged with the probe, filtering out the rest. The device is literally able to “count” the amount of mutated DNA in the sample. The device uses an electrical approach, measuring tiny fluctuations in current as the DNA snakes through a nanopore.

“He’s taken a fundamental biological process of how you move ions across a cell membrane and used that as an engineered detector,” said Michael Hill, interim director of the Dalton Cardiovascular Research Center. “That’s a pretty amazing use of a nano machine.”
While the method is intended to diagnose cancer, it could lead to improved treatments by identifying cancers that are susceptible to treatment by emerging targeted cancer therapeutic agents.
“We’re just entering the medicinal era where that coupling is beginning to occur,” Gates said. “There are medicines targeted at particular mutations in a gene, and that requires rapid and accurate diagnostics to tell you where the tumor has a genetic error in it.”
In the future, the system could be expanded to study genetic markers related to Alzheimer’s Disease and other neurodegenerative conditions, Gu said.
DNA Computing
Researchers at colleges and large tech companies around the world are investigating ways to program biological molecules the way microchips are currently programmed. It’s been proven that books, images and video can be encoded into DNA just as a series of 1s and 0s are encoded into your hard drive.
Were it stored on DNA strands, scientists believe all of the world’s data would total the size of a golf ball, Hill said.
There are several advantages of using DNA from algae, viruses or other biological sources for computing purposes. First, it’s more energy efficient than other data storage systems such as the cloud, Gu said. Additionally, DNA can provide three-dimensional storage whereas circuits are two-dimensional. This increases the data density millions of times.
Once information is imprinted onto a DNA strand, Gu’s nanopore machine could be used to then retrieve that data. The team envisions a DNA “hard drive” or memory in the future, for not only erasable data storage but also high-performance computing.
While DNA computing and data storage is still in its infancy, the interdisciplinary project at Mizzou exemplifies the power of collaboration, Hill said.
“It’s a wonderful blend of chemistry, engineering and biology,” he said. “It’s really exciting to think about all of the applications.”
In addition to Gu and Gates, the research team includes Kevin Gillis, chair and professor of biomedical, biological and chemical engineering.
Be part of world-changing research. Apply today!